With more than 30,000 species, fish is the largest and most ancient group in vertebrates, which also plays important roles in ecosystem and human society. Despite recent progresses in genomics providing information to improve our knowledges in fish, fish genomics falls behind those of birds and mammals, preventing further understandings on evolution, as well as improvements on conservation and applications. Here, in addition to the currently available 215 genomes in 56 orders out of all the 80 orders, we are releasing ten fish genome assemblies representing ten orders. We used an effective assembly strategy thus the released genomes wereof high quality and we have sequenced and are assembling the other 84 species covering 80 families and 36 orders. With experiences in sequencing and assembling those genomes, we are also announcing initiation of Fish 10,000 Genomes Project (Fish10K), in which we aimed at sequencing 10,000 representative fish genomes step by step. In the first and second phases, Fish10K plans to establish high quality reference genomes for at least one species of all orders and all families (along with selected evolutionarily, ecologically or economically important species), using synthetic long reads sequencing, third generation long reads sequencing, along with Hi-C sequencing. Then in the third phase, the rest species will be sequenced using a more sample, cost and time effective way to provide draft genome assemblies. Towards organizing as an open international consortium, Fish10K would like to call for collaborations on resolving important biological questionsusing the generated genome data. Fish10K would also like to recruit global researchers to contribute by providing single or multiple fish species genome data, who can benefit from advance access to all the Fish10K genome data. With previous efforts in fish genomics and the currently initiated Fish10K, we anticipated to accelerate fish genomic researches and ultimately improve our understandingin fish.
The roadmap of Fish10K
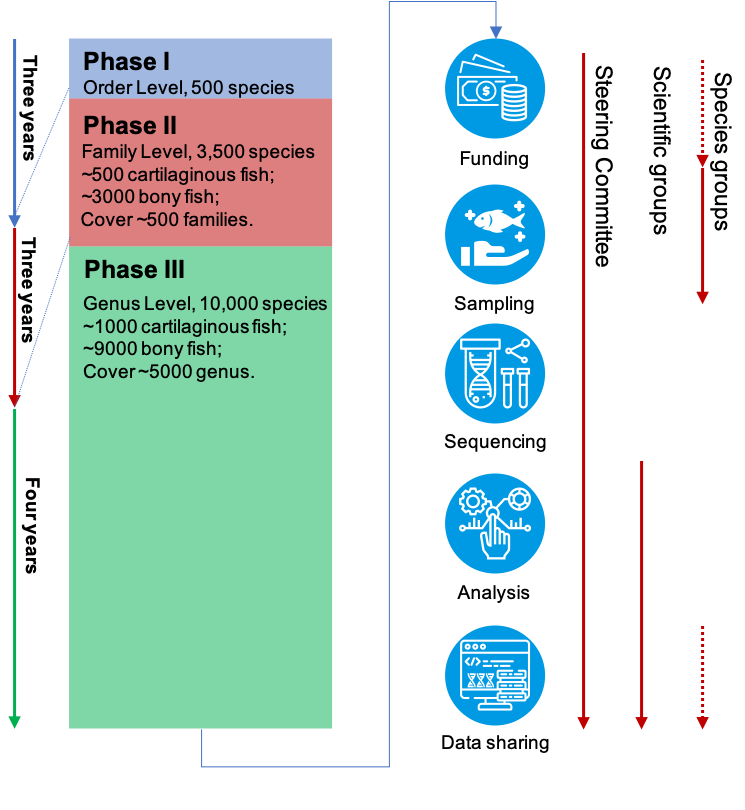
In the first phase, we aimed at sequencing 450 bony fish species and 50 cartilages fish, covered all 80 orders. In the second phase, we would like to sequence about 3,000 more species, covering almost all the 500 families. In the third phase, we will sequence ~6,500 more fish genomes, covering all the ~5,000 genus.
As an important group of species in vertebrates, fish has been studiedin the genome quite comprehensively, with genome sequences of 215 species publicly available, covering 56 orders. Meanwhile, there were about 80 publications on fish genomics in which reference genome sequences were established and analyzed. Those publications focused on phylogeny and evolution of fish (for example, the African coelacanth genome for understanding tetrapod evolution), evolutionary process of specific fish subgroups, genetic mechanisms of adaptation to different environments (Mariana Trench snail fish adapt deep-sea environment and Sinocyclocheilus cavefish for understanding the cave adaptation, etc.) and specific biological processes (Cynoglossus semilaevis genome forunder standing ZW sex chromosome evolution of fish, etc.). However, considering about the abundance of species, as well as theevolutionary and biological questions related to fish, more genome sequences of fish species are required for further comprehensive comparative genomics studies.
With the development of sequencing technology, large-scale genomic studies to construct reference genomes for a group of species and to carry out phylogenomic analysis have become feasible. In vertebrates, Bird 10,000 GenomesProject (B10K) has been initiated after the successful comprehensive phylogenomic study with 45 avian genomes in 2014, aiming at sequencing andassembling all known birds species in three phases. Also following the avian genome project, the Vertebrate Genomes Project (VGP) was launched in 2017 to generate high quality genome assemblies vertebrate species. Similar efforts to construct reference genomes in different groups of species have been made in bats, plants and other species. Those projects accelerated genomic researches indifferent phylogenetic groups, despite they encountered challenges in funding, sampling, sequencing, assembly, as well as data analysis. However, for fish, which makes up more than half of vertebrate species, there was no previous efforts to focus on constructing fish genomes, comparing to B10K for birds. The only large-scale genomic study to our knowledge was Fish-T1K, which sequenced transcriptomes of ray-finned fishes launched in 2013 and finished in 2018. Accelerating fish genomics by large-scale genome sequencing effort, would undoubtedly posed agreat challenge to the research of biodiversity and the utilization and exploitation of fish resources. Under such background, we are initiating Fish 10,000 Genome Project (F10K) here, to sample, sequence, assemble and analyze genomes of 10,000 fish species. We are proposing an effective workflow, in which major challenges over large-scale genomics were considered, to construct high quality reference genomes for as many fish species as possible. Combining all the generated fish genome data together, with developing of effective analyzing methods, we will able to address a series of evolutionary and biological questions related to fish. In order to prove the efficiency of our workflow and the feasibility of this large-scale genome project, we are releasing ten well-assembled genomes from the current genomes we are working on. We hope the released genomes, along with the other 9,990 fish genomes which will be finished within F10K, will be valuable resources for fish researches in near future.