The sampling and sequencing progress of Fish10K
According to FishBase and “Fishes of the world (Fifth Edition)”, we estimated that there were 34,115 fish species, from ~5,000 genus, ~529 families and ~80 orders. These species were mainly divided into 3 class (Elasmobranchii, Myxini, Actinopterygii, Cephalaspidomorphi, Sarcopterygii and Holocephali), in which Elasmobranchii and Holocephali belong to Chondrichthyes; Actinopterygiiand Sarcopterygii belong to Osteichthyes. As mentioned above, there were reference genomes available for at least one species of 56 orders, while for the rest of the orders, reference genomes were required. In addition to that, there are fish orders with large number of species (e.g. Perciformes has 62 families; Siluriformes has 40 families; Scorpaeniformes has 39 families), more reference genomes were required to represent the diverse biological characteristics.
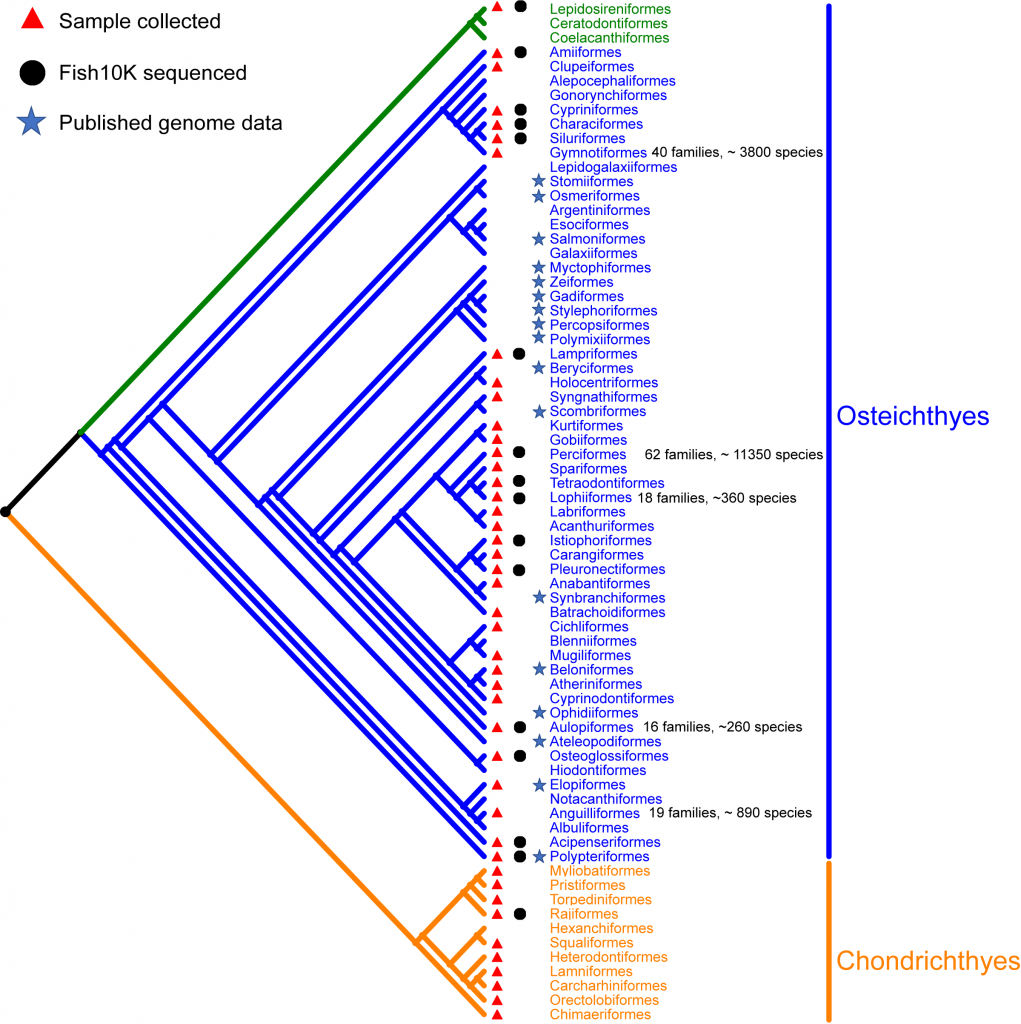
Phylogenetics tree of fish divided into two subgroups: Chondrichthyes (orange) and Osteichthyes (blue and green). The bony fish part consists of two subgroups (The green indicated Sarcopterygii and the blue indicated Actinopterygii). The number offamilies and species of top five largest orders are labeled. Remaining ten orders of bony fish (Caproiformes, Callionymiformes, Gobiesociformes, Icosteiformes, Lepisosteiformes, Moroniformes, Scombrolabraciformes, Scorpaeniformes, Trachichthyiformes and Trachiniformes) and two orders of cartilaginous fish (Rhinopristiformes and Squatiniformes) are not included in the phylogenetic tree, due to their uncertain position.
Assembly information of the publicly available fish genomes
The existing fishes are mainly divided into 3 classes, among which Actinopterygii are all bony fishes, whose number of order and species accounts for more than half of all fishes.
| Species | Taxid | Assembly level | Total sequence length (bp) | Number of scaffolds | Scaffold N50 (bp) | Number of contigs | Contig N50 (bp) | Accession Number |
| Acanthochaenus luetkenii | 473344 | Scaffold | 545759480 | 91087 | 8444 | 131532 | 5636 | GCA_900312575.1 |
| Acanthochromis polyacanthus | 80966 | Scaffold | 991584656 | 30414 | 334400 | 159493 | 16099 | GCF_002109545.1 |
| Acipenser ruthenus | 7906 | Scaffold | 1732545901 | 215913 | 219750 | 446905 | 24377 | GCA_004119895.1 |
| Ageneiosus marmoratus | 2066578 | Scaffold | 1030000983 | 16063 | 223139 | 169048 | 7741 | GCA_003347165.1 |
| Amphilophus citrinellus | 61819 | Scaffold | 844902565 | 6637 | 1216136 | 67543 | 23456 | GCA_000751415.1 |
| Amphiprion ocellaris | 80972 | Scaffold | 880720895 | 6405 | 401715 | 7803 | 324210 | GCF_002776465.1 |
| Amphiprion percula | 161767 | Chromosome | 908955932 | 366 | 38416550 | 1048 | 3123421 | GCA_003047355.2 |
| Anabarilius grahami | 495550 | Scaffold | 991887266 | 80398 | 4459447 | 131192 | 36058 | GCA_003731715.1 |
| Anarrhichthys ocellatus | 433405 | Scaffold | 612774492 | 10816 | 5717598 | 33924 | 41757 | GCA_004355925.1 |
| Anguilla anguilla | 7936 | Scaffold | 1018701900 | 501148 | 59657 | 865467 | 2544 | GCA_000695075.1 |
| Anguilla japonica | 7937 | Scaffold | 966917315 | 83292 | 36264158 | 276102 | 11014 | GCA_003597225.1 |
| Anguilla rostrata | 7938 | Scaffold | 1413032609 | 79209 | 86641 | 307316 | 7355 | GCA_001606085.1 |
| Anoplogaster cornuta | 88656 | Scaffold | 404844788 | 108625 | 4562 | 128159 | 3719 | GCA_900683385.1 |
| Anoplopoma fimbria | 229290 | Contig | 699326415 | 208506 | 5156 | GCA_000499045.1 | ||
| Antennarius striatus | 241820 | Scaffold | 441856641 | 70398 | 9774 | 103068 | 6086 | GCA_900303275.1 |
| Aphyosemion australe | 52653 | Scaffold | 868348903 | 12236 | 1435212 | 30782 | 119465 | GCA_006937985.1 |
| Arapaima gigas | 113544 | Scaffold | 667351951 | 60052 | 285171 | 70752 | 64954 | GCA_007844225.1 |
| Archocentrus centrarchus | 63155 | Chromosome | 932930362 | 188 | 35590001 | 926 | 2146538 | GCF_007364275.1 |
| Arctogadus glacialis | 185735 | Scaffold | 428791846 | 139389 | 3702 | 151920 | 3282 | GCA_900303235.1 |
| Astatotilapia burtoni | 8153 | Scaffold | 831411547 | 8001 | 1194190 | 69074 | 21886 | GCF_000239415.1 |
| Astatotilapia calliptera | 8154 | Chromosome | 880445564 | 249 | 38669361 | 739 | 4438245 | GCF_900246225.1 |
| Astyanax mexicanus | 7994 | Chromosome | 1335239194 | 2415 | 35377769 | 3030 | 1767240 | GCF_000372685.2 |
| Austrofundulus limnaeus | 52670 | Scaffold | 866963281 | 29785 | 1098383 | 168369 | 8097 | GCF_001266775.1 |
| Bathygadus melanobranchus | 630650 | Scaffold | 431202967 | 92290 | 6483 | 112630 | 4956 | GCA_900302375.1 |
| Benthosema glaciale | 125796 | Scaffold | 676314385 | 143923 | 6111 | 188319 | 4393 | GCA_900323375.1 |
| Beryx splendens | 88663 | Scaffold | 533267752 | 117400 | 5987 | 151933 | 4286 | GCA_900312565.1 |
| Betta splendens | 158456 | Chromosome | 441388503 | 70 | 20129463 | 398 | 2497747 | GCF_900634795.2 |
| Boleophthalmus pectinirostris | 150288 | Scaffold | 955752150 | 16620 | 2375582 | 108947 | 20437 | GCF_000788275.1 |
| Boreogadus saida | 44932 | Scaffold | 412070465 | 137701 | 3572 | 147911 | 3221 | GCA_900302515.1 |
| Borostomias antarcticus | 473354 | Scaffold | 430362827 | 104734 | 5368 | 133045 | 3928 | GCA_900323325.1 |
| Bregmaceros cantori | 630652 | Scaffold | 1144104325 | 258566 | 5922 | 319170 | 4452 | GCA_900302395.1 |
| Brosme brosme | 81638 | Scaffold | 412731310 | 114910 | 4650 | 136096 | 3682 | GCA_900302425.1 |
| Brotula barbata | 432164 | Scaffold | 485061200 | 29854 | 45752 | 59402 | 17578 | GCA_900303265.1 |
| Callopanchax toddi | 60409 | Scaffold | 853385936 | 9724 | 1656350 | 45972 | 52012 | GCA_006937965.1 |
| Carapus acus | 1491482 | Scaffold | 387834307 | 46699 | 16922 | 70747 | 9554 | GCA_900312935.1 |
| Carassius auratus | 7957 | Chromosome | 1820635050 | 6216 | 22763433 | 8463 | 821153 | GCF_003368295.1 |
| Coryphaenoides rupestris | 163118 | Scaffold | 829208733 | 47680 | 159738 | 82633 | 20848 | GCA_002895965.1 |
| Cottoperca gobio | 56716 | Chromosome | 609391784 | 322 | 25156145 | 766 | 6330900 | GCF_900634415.1 |
| Cottus rhenanus | 446433 | Scaffold | 563609416 | 164693 | 7249 | 490620 | 2129 | GCA_001455555.1 |
| Cynoglossus semilaevis | 244447 | Chromosome | 470199494 | 31181 | 509861 | 62912 | 27008 | GCF_000523025.1 |
| Cyprinodon variegatus | 28743 | Scaffold | 1035184475 | 9259 | 835301 | 110959 | 20803 | GCF_000732505.1 |
| Cyprinus carpio | 7962 | Chromosome | 1713658011 | 9378 | 7828959 | 53088 | 75080 | GCF_000951615.1 |
| Cyttopsis rosea | 1176755 | Scaffold | 546506150 | 111974 | 7082 | 150231 | 4843 | GCA_900302355.1 |
| Danio rerio | 7955 | Chromosome | 1373454788 | 1917 | 7379053 | 19725 | 1422317 | GCF_000002035.6 |
| Danionella dracula | 623740 | Scaffold | 665208374 | 996 | 10287669 | 1611 | 2300191 | GCA_900490495.1 |
| Danionella translucida | 623744 | Scaffold | 735303417 | 27639 | 340819 | 36005 | 133131 | GCA_007224835.1 |
| Denticeps clupeoides | 299321 | Chromosome | 567401054 | 460 | 22793177 | 924 | 3059612 | GCF_900700375.1 |
| Dicentrarchus labrax | 13489 | Scaffold | 675917103 | 25 | 26439989 | 37781 | 54134 | GCA_000689215.1 |
| Diretmoides pauciradiatus | 1415272 | Scaffold | 672603014 | 147686 | 6032 | 204934 | 4028 | GCA_900660315.1 |
| Diretmus argenteus | 88682 | Scaffold | 302363458 | 107968 | 3367 | 162384 | 2172 | GCA_900660295.1 |
| Echeneis naucrates | 173247 | Chromosome | 544229245 | 38 | 23287306 | 178 | 12371513 | GCF_900963305.1 |
| Electrophorus electricus | 8005 | Scaffold | 551880868 | 8786 | 613956 | 47652 | 37141 | GCF_003665695.1 |
| Epinephelus lanceolatus | 310571 | Chromosome | 1087399367 | 4200 | 46227939 | 22225 | 159800 | GCA_005281545.1 |
| Erpetoichthys calabaricus | 27687 | Chromosome | 3811038701 | 1885 | 199226436 | 7498 | 1143051 | GCF_900747795.1 |
| Esox lucius | 8010 | Chromosome | 940906975 | 811 | 37550661 | 1395 | 3396779 | GCF_004634155.1 |
| Fundulus heteroclitus | 8078 | Scaffold | 1021898560 | 10180 | 1252252 | 120723 | 16688 | GCF_000826765.1 |
| Gadiculus argenteus | 185737 | Scaffold | 396767394 | 123363 | 3951 | 137724 | 3379 | GCA_900302595.1 |
| Gadus chalcogrammus | 1042646 | Scaffold | 448868398 | 130159 | 4335 | 149286 | 3603 | GCA_900302575.1 |
| Gambusia affinis | 33528 | Scaffold | 598663367 | 2943 | 6651460 | 73682 | 17511 | GCA_003097735.1 |
| Gasterosteus aculeatus | 69293 | Scaffold | 467452432 | 10242 | 3715221 | 32646 | 38090 | GCA_006229165.1 |
| Gephyroberyx darwinii | 334984 | Scaffold | 535045983 | 55277 | 18145 | 102041 | 8514 | GCA_900660455.1 |
| Gouania willdenowi | 441366 | Chromosome | 937150793 | 441 | 38978045 | 1594 | 1838341 | GCF_900634775.1 |
| Guentherus altivela | 1263181 | Scaffold | 539598795 | 189411 | 3201 | 205243 | 2928 | GCA_900312595.1 |
| Haplochromis nyererei | 303518 | Scaffold | 830133247 | 7236 | 2525540 | 68053 | 22622 | GCF_000239375.1 |
| Hippocampus comes | 109280 | Scaffold | 493775940 | 37377 | 2034572 | 60478 | 39546 | GCF_001891065.1 |
| Holocentrus rufus | 722565 | Scaffold | 649757301 | 58113 | 21389 | 113697 | 9243 | GCA_900302615.1 |
| Hoplostethus atlanticus | 96778 | Scaffold | 520173038 | 58279 | 15562 | 103281 | 7812 | GCA_900660355.1 |
| Hucho hucho | 62062 | Scaffold | 2487549814 | 71639 | 287338 | 221746 | 37639 | GCA_003317085.1 |
| Hypophthalmichthys molitrix | 13095 | Scaffold | 1104676189 | 107095 | 314181 | 1528765 | 2130 | GCA_004764525.1 |
| Hypophthalmichthys nobilis | 7965 | Scaffold | 1012063666 | 121326 | 83012 | 742098 | 4840 | GCA_004193235.1 |
| Hypoplectrus puella | 146810 | Scaffold | 612290098 | 14375 | 24210077 | 64238 | 22581 | GCA_900610375.1 |
| Ictalurus punctatus | 7998 | Chromosome | 1002389428 | 3163 | 26676597 | 5816 | 2695784 | GCA_004006655.2 |
| Kryptolebias hermaphroditus | 1747188 | Chromosome | 683986837 | 5211 | 27459464 | 32872 | 46411 | GCA_007896545.1 |
| Kryptolebias marmoratus | 37003 | Chromosome | 1002389428 | 3163 | 26676597 | 5816 | 2695784 | GCA_004006655.2 |
| Labeo rohita | 84645 | Scaffold | 1484730970 | 13623 | 1959535 | 42076 | 522833 | GCA_004120215.1 |
| Labeotropheus fuelleborni | 57307 | Scaffold | 70858381 | 58245 | 1204 | 81167 | 1070 | GCA_000150875.1 |
| Labrus bergylta | 56723 | Scaffold | 805480521 | 13466 | 794648 | 13723 | 703847 | GCF_900080235.1 |
| Laemonema laureysi | 1784819 | Scaffold | 306494646 | 86525 | 4715 | 108340 | 3431 | GCA_900303225.1 |
| Lampris guttatus | 81370 | Scaffold | 849277706 | 208230 | 5222 | 253854 | 4051 | GCA_900302545.1 |
| Lamprogrammus exutus | 1592065 | Scaffold | 492850272 | 120937 | 5470 | 145536 | 4213 | GCA_900312555.1 |
| Larimichthys crocea | 215358 | Chromosome | 657939657 | 9998 | 27037660 | 16979 | 277487 | GCF_000972845.2 |
| Lates calcarifer | 8187 | Scaffold | 668481366 | 3808 | 1191366 | 3918 | 1066117 | GCF_001640805.1 |
| Lepisosteus oculatus | 7918 | Chromosome | 945878036 | 2106 | 6928108 | 45200 | 68337 | GCF_000242695.1 |
| Lesueurigobius sanzi | 1365564 | Scaffold | 810626388 | 130360 | 11480 | 186717 | 6729 | GCA_900303255.1 |
| Leuciscus waleckii | 155063 | Scaffold | 752538629 | 4888 | 21959719 | 38277 | 38877 | GCA_900092035.1 |
| Liparis tanakae | 230148 | Scaffold | 498979456 | 27878 | 375216 | 97972 | 9903 | GCA_006348945.1 |
| Lota lota | 69944 | Scaffold | 397499185 | 106616 | 4892 | 128281 | 3803 | GCA_900302385.1 |
| Maccullochella peelii | 135761 | Scaffold | 633241041 | 18198 | 109974 | 31008 | 70439 | GCA_002120245.1 |
| Macquaria australasica | 135765 | Scaffold | 675976139 | 2962 | 845515 | 3369 | 678975 | GCA_005408345.1 |
| Macrourus berglax | 473319 | Scaffold | 399875629 | 118318 | 4291 | 142500 | 3353 | GCA_900302365.1 |
| Malacocephalus occidentalis | 630739 | Scaffold | 350339566 | 95829 | 4932 | 116459 | 3697 | GCA_900312585.1 |
| Mastacembelus armatus | 205130 | Chromosome | 591935101 | 122 | 25090313 | 358 | 8014513 | GCA_900324485.2 |
| Maylandia zebra | 106582 | Chromosome | 957485262 | 1690 | 32660920 | 2331 | 1407748 | GCF_000238955.4 |
| Mchenga conophoros | 35575 | Scaffold | 73425564 | 61923 | 1329 | 85821 | 1066 | GCA_000150855.1 |
| Melanochromis auratus | 27751 | Scaffold | 68238634 | 63297 | 1063 | 86145 | 977 | GCA_000150895.1 |
| Melanogrammus aeglefinus | 8056 | Scaffold | 652790733 | 8420 | 209126 | 15188 | 77605 | GCA_900291075.1 |
| Melanonus zugmayeri | 181410 | Scaffold | 432902915 | 82409 | 7633 | 123515 | 4562 | GCA_900302345.1 |
| Merlangius merlangus | 8058 | Scaffold | 423942190 | 122642 | 4444 | 144924 | 3538 | GCA_900323355.1 |
| Merluccius capensis | 89947 | Scaffold | 414317329 | 110925 | 4774 | 131629 | 3792 | GCA_900312945.1 |
| Merluccius merluccius | 8063 | Scaffold | 401034705 | 102914 | 5117 | 133682 | 3670 | GCA_900312545.1 |
| Merluccius polli | 89951 | Scaffold | 401149128 | 113894 | 4482 | 138586 | 3471 | GCA_900312625.1 |
| Micropterus floridanus | 225391 | Contig | 1001521525 | 249768 | 10978 | GCA_002592385.1 | ||
| Miichthys miiuy | 240162 | Scaffold | 619300777 | 6294 | 1145539 | 20386 | 81271 | GCA_001593715.1 |
| Mola mola | 94237 | Scaffold | 639451992 | 5552 | 8766736 | 51826 | 23239 | GCA_001698575.1 |
| Molva molva | 163112 | Scaffold | 437480619 | 111875 | 5266 | 133189 | 4136 | GCA_900323295.1 |
| Monocentris japonica | 181435 | Scaffold | 556023515 | 52108 | 18672 | 109034 | 8046 | GCA_900323365.1 |
| Mora moro | 248765 | Scaffold | 344961111 | 100621 | 4433 | 125652 | 3267 | GCA_900303205.1 |
| Morone chrysops | 46259 | Scaffold | 620984155 | 84096 | 51932 | 111517 | 20318 | GCA_003610055.1 |
| Morone saxatilis | 34816 | Scaffold | 598109547 | 629 | 25942274 | 70506 | 17211 | GCA_004916995.1 |
| Muraenolepis marmorata | 487677 | Scaffold | 416390766 | 138928 | 3555 | 152594 | 3126 | GCA_900302325.1 |
| Myoxocephalus scorpius | 8097 | Scaffold | 520316443 | 85863 | 9473 | 127441 | 5716 | GCA_900312955.1 |
| Myripristis jacobus | 371672 | Scaffold | 720396841 | 64974 | 21306 | 121390 | 9816 | GCA_900302555.1 |
| Myripristis murdjan | 586833 | Chromosome | 835254674 | 87 | 34950760 | 340 | 14475636 | GCF_902150065.1 |
| Neogobius melanostomus | 47308 | Contig | 1003738541 | 1364 | 2817412 | GCA_007210695.1 | ||
| Neolamprologus brichardi | 32507 | Scaffold | 847910432 | 9099 | 4430025 | 118197 | 13047 | GCF_000239395.1 |
| Nibea albiflora | 240163 | Scaffold | 574466150 | 11977 | 2154052 | 34769 | 55145 | GCA_900327885.1 |
| Nothobranchius furzeri | 105023 | Chromosome | 1242518059 | 6013 | 15858201 | 74941 | 19950 | GCF_001465895.1 |
| Nothobranchius kuhntae | 321403 | Scaffold | 1122656415 | 34756 | 1178460 | 101919 | 22234 | GCA_006942095.1 |
| Notothenia coriiceps | 8208 | Scaffold | 636613682 | 38657 | 217655 | 72571 | 17492 | GCF_000735185.1 |
| Oncorhynchus kisutch | 8019 | Chromosome | 2369932239 | 22813 | 1266128 | 97074 | 58118 | GCF_002021735.1 |
| Oncorhynchus mykiss | 8022 | Chromosome | 2178999613 | 139800 | 1670138 | 559855 | 13827 | GCF_002163495.1 |
| Oncorhynchus nerka | 8023 | Chromosome | 1927141915 | 38027 | 1058586 | 57813 | 329583 | GCF_006149115.1 |
| Oncorhynchus tshawytscha | 74940 | Chromosome | 2425713975 | 15946 | 1728323 | 69485 | 133169 | GCF_002872995.1 |
| Ophiodon elongatus | 225387 | Scaffold | 635567917 | 18379 | 5092707 | 52084 | 31240 | GCA_004358465.1 |
| Oplegnathus fasciatus | 163134 | Scaffold | 766301214 | 4149 | 1126915 | 66839 | 29992 | GCA_003416845.1 |
| Opsanus beta | 95145 | Scaffold | 1028783780 | 345629 | 3335 | 371656 | 3062 | GCA_900660325.1 |
| Oreochromis aureus | 47969 | Scaffold | 918937175 | 12951 | 1102239 | 61878 | 60340 | GCA_005870065.1 |
| Oreochromis niloticus | 8128 | Chromosome | 1005681550 | 2460 | 38839487 | 3010 | 2923640 | GCF_001858045.2 |
| Oreochromis spilurus | 64544 | Contig | 764974731 | 221829 | 11851 | GCA_008269305.1 | ||
| Oryzias javanicus | 123683 | Chromosome | 809679899 | 254 | 35390520 | 751 | 3558013 | GCA_003999625.1 |
| Oryzias latipes | 8090 | Chromosome | 734057086 | 25 | 31218526 | 516 | 2530934 | GCF_002234675.1 |
| Oryzias melastigma | 30732 | Scaffold | 779469774 | 8603 | 23737187 | 56275 | 30057 | GCF_002922805.1 |
| Osmerus eperlanus | 29151 | Scaffold | 342758722 | 73274 | 6820 | 99348 | 4524 | GCA_900302275.1 |
| Oxygymnocypris stewartii | 361644 | Scaffold | 1849224471 | 26281 | 257093 | 26283 | 257093 | GCA_003573665.1 |
| Pachypanchax playfairii | 52664 | Scaffold | 669774067 | 4488 | 3173794 | 19608 | 76641 | GCA_006937955.1 |
| Pagrus major | 143350 | Scaffold | 875465402 | 886260 | 4644 | 1164424 | 2822 | GCA_002897255.1 |
| Pampus argenteus | 206143 | Scaffold | 350448509 | 298139 | 1586 | 532813 | 1001 | GCA_000697985.1 |
| Pangasianodon hypophthalmus | 310915 | Scaffold | 715760110 | 567 | 14288580 | 23339 | 62522 | GCF_003671635.1 |
| Parablennius parvicornis | 171872 | Scaffold | 599249148 | 66539 | 16796 | 129811 | 7343 | GCA_900302745.1 |
| Paralichthys olivaceus | 8255 | Chromosome | 545775252 | 7202 | 3817360 | 38614 | 30544 | GCA_001904815.2 |
| Parambassis ranga | 210632 | Chromosome | 551012959 | 156 | 22993012 | 1677 | 5080925 | GCF_900634625.1 |
| Paramormyrops kingsleyae | 1676925 | Scaffold | 799421083 | 4667 | 1731158 | 47999 | 37656 | GCF_002872115.1 |
| Parasudis fraserbrunneri | 1784818 | Scaffold | 707987062 | 156389 | 6391 | 213462 | 4177 | GCA_900302295.1 |
| Perca flavescens | 8167 | Chromosome | 877456336 | 268 | 37412490 | 1097 | 4268950 | GCF_004354835.1 |
| Perca fluviatilis | 8168 | Scaffold | 958225486 | 31105 | 6260519 | 100821 | 18196 | GCA_003412525.1 |
| Percopsis transmontana | 143327 | Scaffold | 458089168 | 53197 | 15180 | 88141 | 8161 | GCA_900302285.1 |
| Periophthalmodon schlosseri | 1365757 | Scaffold | 679761122 | 46662 | 39308 | 85749 | 16946 | GCA_000787095.1 |
| Periophthalmus magnuspinnatus | 409849 | Scaffold | 701696780 | 26060 | 296161 | 76770 | 28254 | GCA_000787105.1 |
| Phycis blennoides | 163115 | Scaffold | 416766999 | 62684 | 10640 | 132164 | 4532 | GCA_900302315.1 |
| Phycis phycis | 349666 | Scaffold | 346335180 | 100771 | 4502 | 120694 | 3458 | GCA_900302335.1 |
| Pimephales promelas | 90988 | Scaffold | 1219326373 | 73057 | 60380 | 215176 | 7468 | GCA_000700825.1 |
| Planiliza haematocheila | 370040 | Contig | 747342729 | 1453 | 3973280 | GCA_005024645.1 | ||
| Poecilia formosa | 48698 | Scaffold | 748923461 | 3985 | 1574226 | 31058 | 57472 | GCF_000485575.1 |
| Poecilia latipinna | 48699 | Scaffold | 815144743 | 17988 | 279200 | 54625 | 33278 | GCF_001443285.1 |
| Poecilia mexicana | 48701 | Scaffold | 801711499 | 18105 | 275316 | 50601 | 39840 | GCF_001443325.1 |
| Poecilia reticulata | 8081 | Chromosome | 731622281 | 3029 | 5270359 | 40144 | 41908 | GCF_000633615.1 |
| Pollachius virens | 8060 | Scaffold | 394927939 | 116705 | 4344 | 137332 | 3457 | GCA_900312635.1 |
| Polymixia japonica | 81385 | Scaffold | 554895936 | 92198 | 9571 | 134725 | 5803 | GCA_900302305.1 |
| Poropuntius huangchuchieni | 357532 | Scaffold | 760177161 | 625277 | 2931 | 821804 | 2273 | GCA_004124795.1 |
| Pseudochromis fuscus | 280673 | Scaffold | 657041210 | 52042 | 24689 | 91797 | 12029 | GCA_900323345.1 |
| Pseudopleuronectes yokohamae | 245875 | Contig | 547831023 | 525502 | 1994 | GCA_000787555.1 | ||
| Pungitius pungitius | 134920 | Scaffold | 441089565 | 7847 | 302682 | 49352 | 14136 | GCA_003399555.1 |
| Pygocentrus nattereri | 42514 | Scaffold | 1285352492 | 283518 | 1440044 | 325620 | 57732 | GCF_001682695.1 |
| Regalecus glesne | 81389 | Scaffold | 656003707 | 105196 | 9773 | 141447 | 6781 | GCA_900302585.1 |
| Reinhardtius hippoglossoides | 111784 | Scaffold | 677540803 | 4453 | 17640195 | 5488 | 775256 | GCA_006182925.2 |
| Rhamphochromis esox | 163638 | Scaffold | 71295074 | 55751 | 1324 | 78130 | 1126 | GCA_000150935.1 |
| Rondeletia loricata | 88713 | Scaffold | 568597941 | 103827 | 7469 | 140955 | 5112 | GCA_900302605.1 |
| Salarias fasciatus | 181472 | Chromosome | 797507141 | 203 | 32729575 | 805 | 2597836 | GCF_902148845.1 |
| Salmo salar | 8030 | Chromosome | 2966890203 | 241573 | 1366254 | 368060 | 57618 | GCF_000233375.1 |
| Salmo trutta | 8032 | Chromosome | 2371880186 | 1441 | 52209666 | 5378 | 1703178 | GCF_901001165.1 |
| Salvelinus alpinus | 8036 | Chromosome | 2169553147 | 16702 | 1018695 | 97014 | 55619 | GCF_002910315.2 |
| Sander lucioperca | 283035 | Scaffold | 900461225 | 1312 | 4929547 | 1347 | 4695595 | GCA_008315115.1 |
| Sardina pilchardus | 27697 | Scaffold | 949617276 | 117259 | 96617 | 194510 | 9398 | GCA_900499035.1 |
| Scartelaos histophorus | 166764 | Scaffold | 695008792 | 156044 | 15105 | 209353 | 8806 | GCA_000787155.1 |
| Scleropages formosus | 113540 | Chromosome | 784563014 | 72 | 31084684 | 217 | 9102216 | GCF_900964775.1 |
| Scophthalmus maximus | 52904 | Chromosome | 524979463 | 22 | 24811384 | 21326 | 54836 | GCA_003186165.1 |
| Sebastes aleutianus | 214485 | Scaffold | 899650391 | 10489 | 340062 | 110635 | 10838 | GCA_001910805.2 |
| Sebastes koreanus | 290523 | Contig | 725092264 | 147157 | 16662 | GCA_004335335.1 | ||
| Sebastes minor | 214483 | Scaffold | 681652711 | 166448 | 7676 | 812852 | 1901 | GCA_001910765.2 |
| Sebastes nigrocinctus | 72089 | Scaffold | 746044620 | 15872 | 116274 | 89356 | 13471 | GCA_000475235.3 |
| Sebastes norvegicus | 394699 | Scaffold | 717740616 | 75627 | 16564 | 117709 | 9467 | GCA_900302655.1 |
| Sebastes nudus | 1617787 | Contig | 724045237 | 180312 | 11290 | GCA_004335365.1 | ||
| Sebastes rubrivinctus | 72099 | Scaffold | 756296653 | 68206 | 30046 | 136109 | 13541 | GCA_000475215.1 |
| Sebastes schlegelii | 214486 | Contig | 728476695 | 146105 | 14246 | GCA_004335315.1 | ||
| Sebastes steindachneri | 201708 | Scaffold | 648011071 | 279232 | 4288 | 1089366 | 1311 | GCA_001910785.2 |
| Selene dorsalis | 179366 | Scaffold | 528779420 | 36113 | 32464 | 85660 | 11209 | GCA_900303245.1 |
| Seriola dumerili | 41447 | Scaffold | 677686174 | 34656 | 5812906 | 41188 | 249509 | GCF_002260705.1 |
| Seriola lalandi | 302047 | Scaffold | 766364468 | 7606 | 411616 | 63278 | 37711 | GCA_003054885.1 |
| Seriola quinqueradiata | 8161 | Scaffold | 639269536 | 384 | 5610255 | 1312 | 872227 | GCA_002217815.1 |
| Seriola rivoliana | 173321 | Scaffold | 666141578 | 1343 | 9509606 | 3939 | 740108 | GCA_002994505.1 |
| Simochromis diagramma | 43689 | Scaffold | 848827444 | 823 | 8960300 | 1764 | 2231376 | GCA_900408965.1 |
| Sinocyclocheilus anshuiensis | 1608454 | Scaffold | 1632718266 | 85682 | 1284143 | 254423 | 17271 | GCF_001515605.1 |
| Sinocyclocheilus grahami | 75366 | Scaffold | 1750287761 | 31277 | 1156368 | 168074 | 29353 | GCF_001515645.1 |
| Sinocyclocheilus rhinocerous | 307959 | Scaffold | 1655786410 | 164173 | 945738 | 314963 | 18758 | GCF_001515625.1 |
| Sparus aurata | 8175 | Chromosome | 833595063 | 176 | 35791275 | 1224 | 2862625 | GCF_900880675.1 |
| Sphaeramia orbicularis | 375764 | Chromosome | 1342662642 | 340 | 57165184 | 2183 | 2360121 | GCF_902148855.1 |
| Spondyliosoma cantharus | 50595 | Scaffold | 680472139 | 47064 | 28198 | 97735 | 11633 | GCA_900302685.1 |
| Squalius pyrenaicus | 263744 | Contig | 48139320 | 40926 | 1710 | GCA_001403095.1 | ||
| Stegastes partitus | 144197 | Scaffold | 800491834 | 5818 | 411659 | 42060 | 43010 | GCF_000690725.1 |
| Stylephorus chordatus | 409996 | Scaffold | 488488587 | 128468 | 4684 | 170584 | 3373 | GCA_900312615.1 |
| Symphodus melops | 171736 | Scaffold | 533823763 | 50156 | 21275 | 95080 | 9362 | GCA_900323315.1 |
| Syngnathus acus | 161584 | Chromosome | 324331233 | 87 | 14974571 | 130 | 11959915 | GCA_901709675.1 |
| Tachysurus fulvidraco | 1234273 | Scaffold | 713810725 | 663 | 3653474 | 2402 | 980445 | GCF_003724035.1 |
| Takifugu bimaculatus | 433685 | Chromosome | 371675663 | 22 | 16786025 | 1055 | 1398332 | GCA_004026145.1 |
| Takifugu flavidus | 433684 | Chromosome | 366286831 | 867 | 15676631 | 1111 | 4357567 | GCA_003711565.2 |
| Takifugu rubripes | 31033 | Chromosome | 384126662 | 128 | 16705553 | 530 | 3136617 | GCF_901000725.2 |
| Tenualosa ilisha | 373995 | Scaffold | 815647530 | 124209 | 188026 | 131117 | 129889 | GCA_003651195.1 |
| Thalassoma bifasciatum | 76338 | Scaffold | 1095910316 | 379332 | 155821 | 397893 | 122955 | GCA_008086565.1 |
| Thunnus albacares | 8236 | Scaffold | 728212003 | 38995 | 46920 | 84919 | 16808 | GCA_900302625.1 |
| Thunnus orientalis | 8238 | Contig | 684497465 | 133062 | 8235 | GCA_000418415.1 | ||
| Thunnus thynnus | 8237 | Scaffold | 648208697 | 354425 | 3045 | 450338 | 2430 | GCA_003231725.1 |
| Thymallus thymallus | 36185 | Chromosome | 1564834359 | 3831 | 32985317 | 204386 | 31774 | GCA_004348285.1 |
| Trachinotus ovatus | 173339 | Scaffold | 648062395 | 138 | 29494812 | 749 | 1846793 | GCA_900607315.1 |
| Trachyrincus murrayi | 241836 | Scaffold | 452416606 | 40927 | 19998 | 114476 | 6231 | GCA_900323305.1 |
| Trachyrincus scabrus | 562814 | Scaffold | 369861760 | 80958 | 6379 | 119164 | 3900 | GCA_900303215.1 |
| Triplophysa siluroides | 422203 | Scaffold | 583428323 | 1002 | 2872994 | 1039 | 2549348 | GCA_006030095.1 |
| Trisopterus minutus | 80722 | Scaffold | 334717091 | 106116 | 3976 | 122084 | 3248 | GCA_900302415.1 |
| Typhlichthys subterraneus | 940470 | Scaffold | 555559596 | 84841 | 9654 | 106331 | 7314 | GCA_900302405.1 |
| Xiphophorus couchianus | 32473 | Chromosome | 688541509 | 68 | 30550352 | 297 | 15315838 | GCF_001444195.1 |
| Xiphophorus hellerii | 8084 | Chromosome | 733126988 | 85 | 26465357 | 561 | 7119835 | GCA_003331165.1 |
| Zeus faber | 64108 | Scaffold | 610433400 | 135758 | 6332 | 172424 | 4642 | GCA_900323335.1 |
The collected and sequenced species
Among these more than 34,000 species, fewer species has been sequenced. Data mining of sequenced fish from NCBI shows that the sequenced fish are mainly represented a few orders. For some orders with a large number of families (e.g. Perciformes has 62 families; Siluriformes has 40 families; Scorpaeniformes has 39 families), it usually need multiple families of genomes data to reflect the biological characteristics ofthis order. But, until now, most fish still have no genome data, remained the gaps in construction of tree of life in order-level and/or family-level. Through the execution of F10K, these gaps will be filled. In the preliminary work of the project, we have collected 324 fish species, including Chondrichthyes and Osteichthyes,covered 196 family and 68 orders. Until now, 105 fish species have received the high-quality genome assembly.
| Species | Stlfr (Gb) | Hi-C (Gb) | ATAC (Gb) | RNA (Gb) | Nanopore (Gb) |
| Mormyrus caschive Linnaeus | 119.39 | 142.63 | 12.56 | 18.32 | 9.61 |
| Campylomormyrus elephas | 121.14 | 40.10 | 12.38 | preparing | preparing |
| Semaprochilodus insignis | 120.65 | 78.58 | 11.26 | Fail | preparing |
| Panaque nigrolineatus | 118.51 | preparing | 13.78 | 18.14 | 11.33 |
| Glossolepis incisus | 121.70 | preparing | 10.80 | 19.56 | 9.80 |
| Kuhlia marginata | 114.67 | 71.48 | 12.84 | preparing | preparing |
| Distichodus sexfasciatus | 125.09 | preparing | 1.88 | 17.58 | 14.12 |
| Megalops atlanticus | 117.61 | preparing | 9.06 | 22.36 | preparing |
| Nemacheilidae* | 115.22 | 143.26 | preparing | preparing | preparing |
| Helostoma temminckii | 117.99 | 98.46 | 8.98 | Fail | 11.11 |
| Asterophysus batrachus | 319.01 | preparing | preparing | preparing | preparing |
| Callichthyidae* | 114.60 | preparing | preparing | preparing | preparing |
| Sypreparingdontis nigriventris | 114.76 | preparing | preparing | preparing | preparing |
| Sahyadria denisonii | 116.50 | preparing | preparing | preparing | preparing |
| Iriatherina werneri | 123.63 | preparing | preparing | preparing | preparing |
| Pantodon buchholzi | 99.81 | preparing | preparing | preparing | preparing |
| Boulengerella lucia | 122.82 | 68.36 | 11.12 | 22.80 | preparing |
| Crossocheilus oblongus | 122.12 | preparing | preparing | preparing | preparing |
| Kryptopterus bicirrhis | 123.83 | preparing | preparing | preparing | preparing |
| Acestrorhynchus altus | 125.46 | sequencing | 6.50 | 19.40 | 12.06 |
| Mopreparingtremus palembangensis | 121.83 | 82.08 | 17.54 | 18.36 | preparing |
| Tetraodon fluviatilis | 122.42 | sequencing | 10.12 | 19.32 | preparing |
| Hydrocynus vittatus | 92.93 | 56.64 | 5.14 | 21.30 | 12.75 |
| Tetraodon palembangensis | 120.40 | 19.00 | 11.12 | 20.64 | 12.32 |
| Malapterurus electricus | 118.87 | 124.02 | 8.96 | 16.22 | 9.47 |
| Naso vlamingii | 131.47 | sequencing | preparing | 18.86 | 12.13 |
| Centropyge bicolor | 130.47 | 134.61 | preparing | preparing | preparing |
| Synchiropus splendidus | 130.25 | sequencing | preparing | 12.44 | preparing |
| Corythoichthys haematopterus | 364.22 | preparing | preparing | preparing | preparing |
| Diodon holocanthus | 127.52 | sequencing | preparing | 22.86 | 11.66 |
| Chelmon rostratus | 134.64 | 134.22 | preparing | preparing | preparing |
| Auchepreparingglanis occidentalis | 128.79 | 19.24 | 5.82 | 17.84 | 10.15 |
| Heterotis niloticus | 119.30 | 21.24 | 10.06 | 23.02 | 10.46 |
| Crenicichla johanna | 120.14 | 136.03 | 8.56 | 19.88 | 10.79 |
| Hydrolycus armatus | 120.76 | 54.46 | 7.80 | 18.48 | preparing |
| Macrochirichthys macrochirius | 114.39 | 66.38 | 9.72 | 21.38 | 10.27 |
| Hepsetus odoe | 118.61 | 66.86 | 2.68 | 22.26 | 11.73 |
| Salminus brasiliensis | 120.54 | 73.60 | 10.96 | 25.50 | 12.05 |
| Metynnis lippincottianus | 341.02 | preparing | preparing | preparing | 12.33 |
| Polypterus endlicheri | 336.58 | preparing | preparing | preparing | preparing |
| Brachyplatystoma tigrinum | 94.44 | preparing | preparing | preparing | preparing |
| Sypreparingdontis flavitaeniatus | 73.88 | preparing | preparing | preparing | preparing |
| Hexanematichthys leptaspis | 85.30 | preparing | preparing | preparing | preparing |
| Leporinus frederici | 302.09 | preparing | preparing | preparing | preparing |
| Garra orientalis | 99.14 | preparing | preparing | preparing | preparing |
| Gyripreparingcheilus* | 98.65 | 137.06 | 8.36 | 22.88 | 11.19 |
| Aplocheilus lineatus | 97.10 | preparing | preparing | preparing | preparing |
| Phenacogrammus interruptus | 104.41 | preparing | preparing | preparing | preparing |
| Apteropreparingtus albifrons | 108.95 | preparing | preparing | preparing | 10.00 |
| Hoplias aimara | 95.89 | preparing | preparing | preparing | preparing |
| Lethrinus | 126.19 | preparing | preparing | preparing | 11.38 |
| Therapon oxyrhynchus | 124.76 | preparing | preparing | preparing | 18.53 |
| Eleotridae* | 125.74 | preparing | preparing | preparing | 19.64 |
| Datnioides pulcher | 122.38 | preparing | preparing | preparing | 11.24 |
| Rhipreparinggobius giurinus | 133.18 | preparing | preparing | preparing | 11.60 |
| Chaetodon trifasciatus | 128.29 | preparing | preparing | preparing | 10.32 |
| Cobitis sinensis | 130.76 | preparing | preparing | preparing | preparing |
| Eleotris oxycephala | 137.64 | preparing | preparing | preparing | preparing |
| Harpadon nehereus | 131.32 | preparing | preparing | preparing | preparing |
| Sillago japonica | 129.92 | preparing | preparing | preparing | preparing |
| Brachirus orientalis | 137.34 | preparing | preparing | preparing | preparing |
| Apogonidae* | 131.97 | preparing | preparing | preparing | preparing |
| Triacanthus biaculeatus | 134.93 | preparing | preparing | preparing | preparing |
| Anabas testudineus | 130.97 | preparing | preparing | preparing | preparing |
| Lutjanus fulviflamma | 134.98 | preparing | preparing | preparing | preparing |
| Bostrychus sinensis | 135.54 | preparing | preparing | preparing | preparing |
| Urapreparingscopiformes | 134.82 | preparing | preparing | preparing | preparing |
| Arothron hispidus | 133.51 | preparing | preparing | preparing | preparing |
| Abudefduf vaigiensis | 132.93 | preparing | preparing | preparing | preparing |
| Gympreparingthorax | 140.25 | preparing | preparing | preparing | preparing |
| Mopreparingpterus albus | 136.21 | preparing | preparing | preparing | preparing |
| Eleotris oxycephala | 125.41 | preparing | preparing | preparing | preparing |
| Sillaginidae | 115.00 | preparing | preparing | preparing | preparing |
| Gerres acinaces | 118.53 | preparing | preparing | preparing | preparing |
| Platycephalidae | 131.16 | preparing | preparing | preparing | preparing |
| Sillaginidae | 122.42 | preparing | preparing | preparing | preparing |
| Inimicus japonicus | 126.58 | preparing | preparing | preparing | preparing |
| Platycephalidae | 118.44 | preparing | preparing | preparing | preparing |
| Johnius grypotus | 120.79 | preparing | preparing | preparing | preparing |
| Gobiidae | 131.75 | preparing | preparing | preparing | preparing |
| Pisodopreparingphis cancrivorus | 126.37 | preparing | preparing | preparing | preparing |
| Leiognathus brevirostris | 121.58 | preparing | preparing | preparing | preparing |
| Hemibarbus labeo | 130.99 | preparing | preparing | preparing | preparing |
| Zebrasoma veliferum | 139.93 | preparing | preparing | preparing | preparing |
| Siganus unimaculatus | 139.93 | preparing | preparing | preparing | preparing |
| Chaetodon auriga | 138.54 | preparing | preparing | preparing | preparing |
| Carassioides cantonensis | 121.21 | preparing | preparing | preparing | preparing |
| Varicorhinus lepturus | 119.60 | preparing | preparing | preparing | preparing |
| Carassioides cantonensis | 124.74 | preparing | preparing | preparing | preparing |
| Balitora brucei | 96.75 | preparing | preparing | preparing | preparing |
| Opsariichthys bidens | 141.76 | preparing | preparing | preparing | preparing |
| Chapreparingdichthys erythropterus | 141.25 | preparing | preparing | preparing | preparing |